Welcome…

… to the R’n’R lab, where you can learn about DNA Replication and Repair, meet Modesto Redrejo Rodríguez and listen to Rock & Roll music.
Our lab is at the School of Medicine of UAM (Madrid) and it also belongs to the Instituto de Investigaciones Biomédicas “Sols-Morreale”, a joint CSIC/UAM Research Institute on biomedicine. The RnR group is also available at IIB site.

![]()
Lab Scope
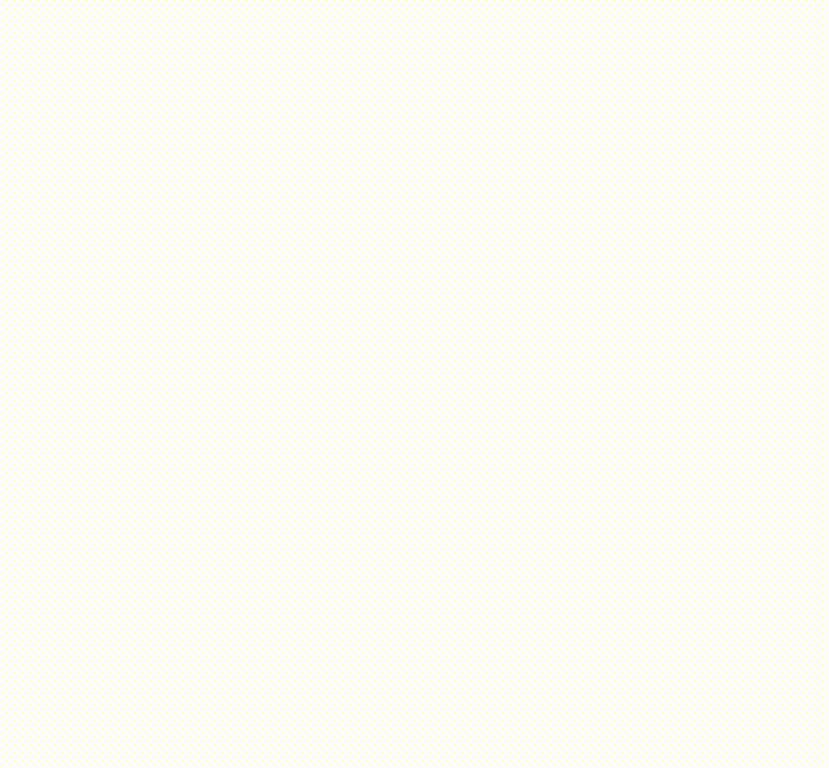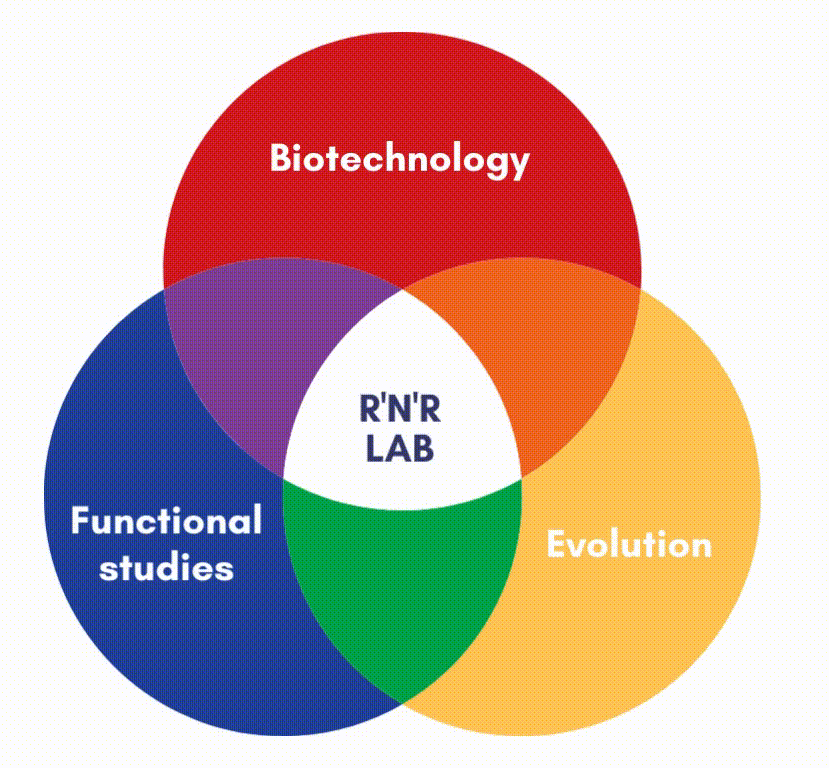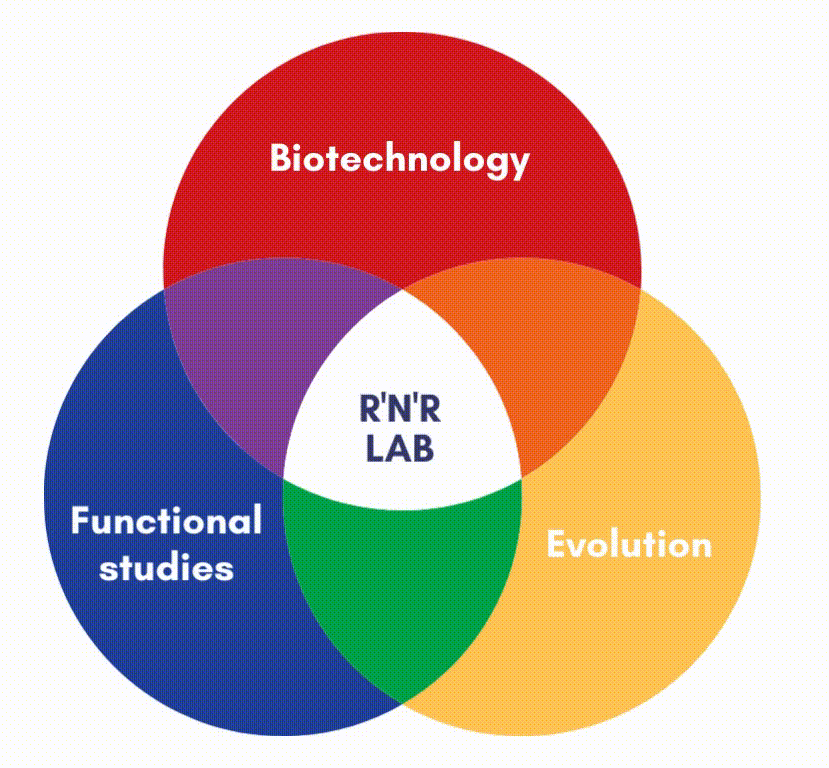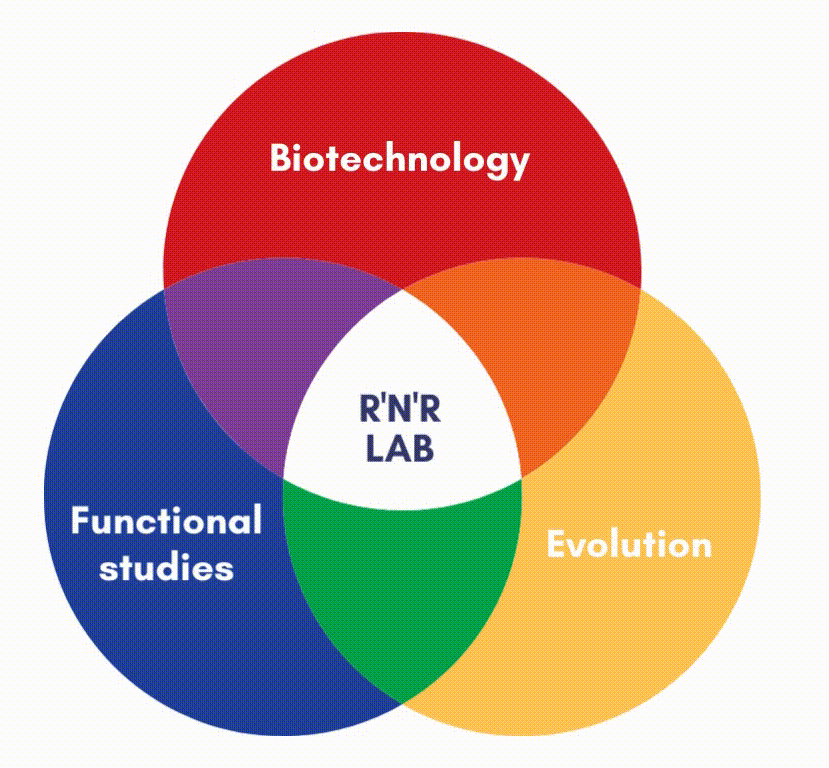
In our lab, we focus on the protein factors (enzymes) that carry out DNA replication and repair.
DNA polymerases are the main responsible for copying the information stored in each DNA strand. We are also interested in other enzymes, such as DNA binding proteins, or DNA Glycosylases, and AP endonuclease, which can surgically identify and remove damage from the double helix. We carry out structure-function studies along with phylogenetic approaches to understand the mechanisms of DNA maintenance and the evolution of those mechanisms.
We use simple genetic models, like transposons, virus and bacteria to analyze the molecular mechanisms that maintain genetic information in the DNA molecule. This also allows us to devise new methods and biotechnological applications.
We are currently focused in DNA polymerases but new projects are being cooked under low heat!!
Phylosophy
We are a small lab that aspires to be a good place to learn and science together. As the great Professor Margarita Salas used to say, our ultimate goal is “to work hard and to work well”. To achieve that, we team up in a collaborative science environment.
We use Benchling as electronic laboratory notebook for daily work. All new students get the following Benchling notes about the lab work:
This is a Quarto website, hosted is on a GitHub repo under CC BY-NC license. To learn more about Quarto see https://quarto.org. You can also check our R (& Rstudio)course in the Teaching section.
Also, it is likely that you find some mistypes or even some big mistakes throughout these site. I will appreciate it if you let me know about anything that could be corrected or just improved.